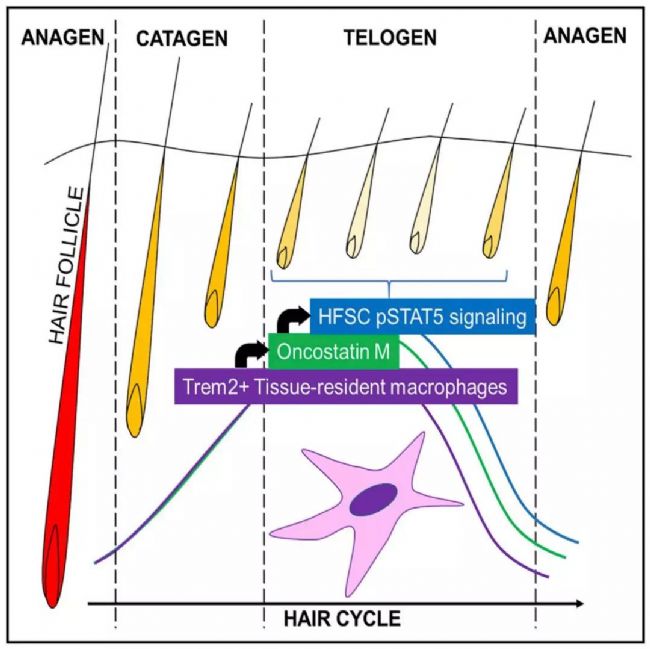
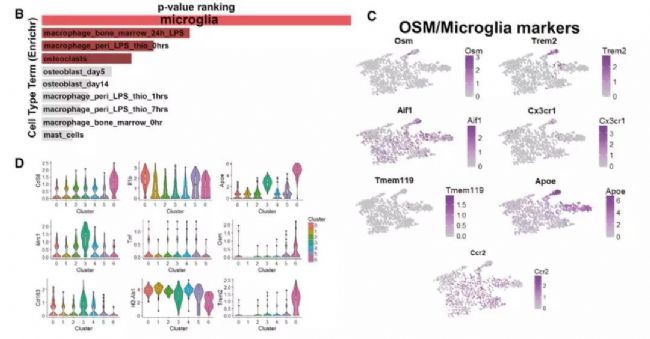
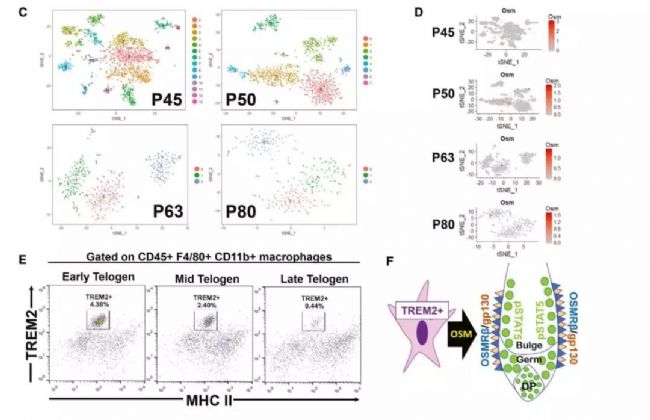
Single-cell transcriptome sequencing is a very popular high-throughput omics research tool in the past two years. However, is it suitable for all studies? Can all problems be explained by this method? This is a question worth exploring. From the existing research reports, single-cell transcriptomics research has solved important biological problems such as cell map and developmental process. But in a relatively traditional research direction, can single-cell transcriptome sequencing methods be used to make research more in-depth and outstanding? In this issue, through the analysis of the key content of the recent literature published in "Cell Stem Cell", to understand the subtle use of single-cell transcriptomics in the study of germinal mechanisms , how to explain the regulatory pathways in the critical period of germinal development Direct evidence is provided. Research Background The mammalian hair life cycle is experienced by the hair follicles (HF) through three important fine-tuning stages represented by the growth phase (anagen), the catagen (catagen) and the telogen (telogen). Keratinocytes derived from hair follicle stem cells (HFSCs), when activated by an activation signal such as Wnt or Shh, promote cell proliferation during the growth phase; when they receive a suppression signal such as BMPs during the resting phase, they promote cell quiescence. The potential upstream regulatory signals of the JAK-STAT pathway that regulate resting quiescence are considered to be factors in some cytokine families. Oncostatin (OSM) has been shown to inhibit the growth phase induced by plucking, while IL-6 can induce hair follicles to enter the degenerative phase from the growth phase. OSM and IL-6 belong to a gp130-dependent family of cytokines that induce the production of LIF, CT-1, and CNTF. In the present study, the researchers found that OSM inhibits the hair growth process induced by the JAK-STAT pathway, possibly as an endogenous statin maintenance quiescent period. Through further single-cell sequencing and validation, it was found that OSM is a process in which a class of TREM2+ macrophages are produced early in the resting phase to affect the hair growth cycle. Figure 1 shows the main results of the study Research subject In order to study the regulation of OSM in hair growth, this study conducted a series of traditional molecular biology experiments, and obtained the following conclusions: So what did the researchers do with single-cell transcriptome sequencing and what did they find? The researchers performed single-cell transcriptome sequencing (10X Genomics platform) on dermal CD45+ immune cells in three periods of late (P45) (P50, P63) late (P80) rest periods. Analysis showed that cells in the P45 period were divided into 13 different clusters. Using existing macrophage markers CD68, F4/80 (Adgre1) and Csf1r, P45 macrophages were subdivided into five different clusters: 0, 1, 2, 5, 11. OSM is mainly expressed in Cluster11 with high abundance. 8. The macrophage producing OSM is TREM2+ and has a unique gene expression profile. By clustering the significantly up-regulated genes in each cluster, it can be found that Cluster 6 cells and the "M2-like" Cluster3 expression patterns are very similar. Interestingly, these two types of cells can be distinguished by the TREM2 marker, which is specifically expressed in Cluster 6 producing OSM. Using the online resource Enrichr to analyze the cluster 6 gene, its function is mainly related to microglia, which is the long-term tissue resident macrophages of the central nervous system. Microglial cell-associated markers (Apoe, Aif-1, Cx3cr1 and Tmem119) are expressed in this class of cells. Therefore, OSM and TREM2 are genes that are highly expressed in culster 6. Figure 3 Expression profile of macrophages producing OSM 9. TREM2+ macrophages are closely related to HFSCs The researchers selected a 30 μm tissue block for mouse dermal tissue for marker immunofluorescence detection. It was found that Aif-1 and CD11b colocalize with OSM in P45 phase hair follicle immune cells; TREM2 and F4/80 and OSM are close to the resting HFSCs. Colocalization in phagocytes. The results in this section indicate that OSM is produced by TREM2+ macrophages in resting HFs. 10. TREM2+ macrophages will decrease after rest period If TREM2+ macrophages inhibit the increase in HFSC, they should be reduced before the next round of growth. To validate this hypothesis, the researchers further analyzed single-cell data at three different time points in the resting period. The results showed that the diversity of CD45+ immune cells gradually decreased with the development of the rest period, from 13 types of P45 to 3 types of P63. When the cell group producing OSM is from P45 to P50, it is also less, and P63 and P80 are further reduced. Such results were also verified by flow cytometry. Trajectory analysis of OSM and microglial markers also showed that their expression continued to decrease during the resting period. Therefore, during the resting period, TREM2+ macrophages affect the OSMRβ-STAT5 signaling pathway by producing OSM, and maintain the quiescent state of HFSC cells. Through the above analysis, the study not only found out how OSM affects the rest period, but also found that the true source of OSM is a class of TREM2+ macrophages. Later, in the resting period, genetically, antibody neutralizing and small molecule inhibitors inhibited macrophages and found that it can effectively induce the growth phase. The above is the main content of the study and the key analysis in the single cell research process. It can be seen that this study, through the single-cell transcriptome experiment, explains on which the specific type of macrophage is produced by OSM. It also discovers the changes of OSM in three different phases of the rest period. Said to subtly use the single cell technology very perfectly. Therefore, single-cell transcriptomics sequencing technology, as long as it can be rationally designed and planned, I believe that the vast majority of research can be used. Magnetic Bead Nucleic Acid Extraction Reagent
The magnetic bead method nucleic acid extraction kit is a high-tech product that combines biological science and nanomaterial science. It is a major breakthrough in my country's nucleic acid extraction and purification technology. .
Magnetic bead method for nucleic acid extraction has incomparable advantages over traditional DNA extraction methods, which are mainly reflected in: 1. It can realize automatic and large-scale operation. At present, there are 96-well nucleic acid automatic extraction instruments, and the extraction time of one sample can be realized. The processing of 96 samples complies with the high-throughput operation requirements of biology, enabling rapid and timely response to infectious disease outbreaks, which makes traditional methods unmatched; â‘¡ The operation is simple and time-consuming, and the entire extraction process has only four steps , most of them can be completed within 36-40 minutes; â‘¢ Safe and non-toxic, no toxic reagents such as benzene and chloroform in traditional methods are used, and the damage to experimental operators is minimized, which fully conforms to modern environmental protection concepts; â‘£ Magnetic beads and nucleic acids The specific binding of the extracted nucleic acid results in high purity and high concentration of the extracted nucleic acid.
According to the same principle as the silica membrane spin column, the superparamagnetic silica nano-magnetic beads are prepared after the surface of the superparamagnetic nanoparticles is modified and modified by nanotechnology. The magnetic beads can specifically recognize and efficiently bind to nucleic acid molecules on a microscopic interface. Using the superparamagnetic properties of silica-coated nanomagnetic microspheres, under the action of Chaotropic salts (guanidine hydrochloride, guanidine isothiocyanate, etc.) and an external magnetic field, samples from blood, animal tissues, food, pathogenic microorganisms, etc. The isolated DNA and RNA can be used in clinical disease diagnosis, blood transfusion safety, forensic identification, environmental microbial testing, food safety testing, molecular biology research and other fields.
Magnetic bead nucleic acid extraction can generally be divided into four steps: lysis-binding-washing-elution.
Genetic testing will surely become a new symbol of the development of the biological industry. The emergence of high-throughput, automated nucleic acid extraction methods will reduce the labor cost of genetic testing, make large-scale testing a reality, and make it possible for genetic testing to reach ordinary people.
Magnetic Bead Nucleic Acid Extraction Reagent,Nucleic Acid Extraction Reagent,Extraction Kit Magnetics Bead Method,Magnetic Bead Acid Nucleic Extraction Kit Jilin Sinoscience Technology Co. LTD , https://www.jlgkscience.com
2. OSMRβ and its co-receptor gp130 co-localize to activate pSTAT5 in HFSCs cell bulge and germ during early and mid-term rest periods;
3. The interference of the OSMRb signal path shortens the second rest period;
4. Interfering with OMSRβ and STAT5 expression during the resting phase promotes the value-added and growth phase of HFSC;
5. OSM in dermal cells is derived from macrophages in resting tissues;
6. The OSM produced by macrophages can inhibit the proliferation of HFSC in vitro;
Through the above experiments, the research has opened up the important regulation process of OSM in the process of hair growth and the main source of OSM, which should be the standard operation of traditional biomolecular mechanism experiments. However, the study did not stop at this conclusion, using the single-cell transcriptome sequencing approach, the study continues to take a big step forward. 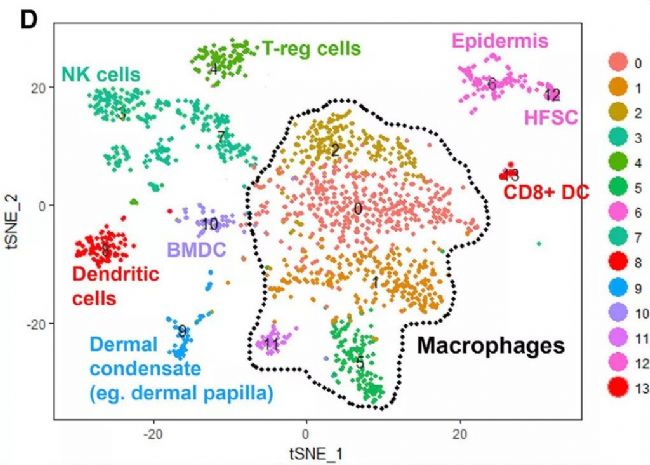
References: Wang, ECE, et al., A Subset of TREM2(+) Dermal Macrophages Secretes Oncostatin M to Maintain Hair Follicle Stem Cell Quiescence and Inhibit Hair Growth. Cell Stem Cell, 2019. 24(4): p. 654- 669 e6.
Click http:// to fill out the inquiry form online and we will contact you as soon as possible.
"Cell Stem Cell" uses single-cell sequencing to analyze the deep-level regulation mechanism of the hair growth process
Figure 2 Single cell sequencing results tSNE cell classification results
For a more detailed classification, the researchers reclassified cells expressed by macrophage markers, resulting in a total of seven new subgroups. These macrophage clusters show relatively uniform CD68 expression but are heterogeneous relative to classical inflammation and anti-inflammatory markers. Among them, Cluster 3 mainly expresses anti-inflammatory marker Mrc2 (CD206) and CD163, while Cluster 0 and 1 mainly express inflammation markers IL-1b and MHC II (H2-Ab1). It is worth noting that OSM is mainly limited in Cluster 6 in the new classification.
Figure 4 Characteristics of immune cells and OSM regulation patterns at different time points